9 Stage Laser-Plasma Accelerator Surrogate
This example models an electron beam accelerated through nine stages of laser-plasma accelerators with ideal plasma lenses providing the focusing between stages. For more details, see:
Sandberg R T, Lehe R, Mitchell C E, Garten M, Qiang J, Vay J-L and Huebl A. Synthesizing Particle-in-Cell Simulations Through Learning and GPU Computing for Hybrid Particle Accelerator Beamlines. Proc. of Platform for Advanced Scientific Computing (PASC’24), submitted, 2024.
Sandberg R T, Lehe R, Mitchell C E, Garten M, Qiang J, Vay J-L and Huebl A. Hybrid Beamline Element ML-Training for Surrogates in the ImpactX Beam-Dynamics Code. 14th International Particle Accelerator Conference (IPAC’23), WEPA101, 2023. DOI:10.18429/JACoW-IPAC2023-WEPA101
A schematic with more information can be seen in the figure below:
![[fig:lpa_schematic] Schematic of the 9 stages of laser-plasma accelerators.](https://user-images.githubusercontent.com/10621396/289956389-c7463b99-fb56-490a-8511-22c43f45cdf8.png)
Fig. 6 [fig:lpa_schematic] Schematic of the 9 stages of laser-plasma accelerators.
The laser-plasma accelerator elements are modeled with neural network surrogates,
previously trained and included in models.
The neural networks require normalized input data; the normalizations can be found in datasets.
We use a 1 GeV electron beam with initial normalized rms emittance of 1 mm-mrad.
In this test, the initial and final values of \(\sigma_x\), \(\sigma_y\), \(\sigma_t\), \(\epsilon_x\), \(\epsilon_y\), and \(\epsilon_t\) must agree with nominal values.
Run
This example can be only be run with Python:
Python script:
python3 run_ml_surrogate.py
For MPI-parallel runs, prefix these lines with mpiexec -n 4 ... or srun -n 4 ..., depending on the system.
#!/usr/bin/env python3
#
# Copyright 2022-2023 ImpactX contributors
# Authors: Ryan Sandberg, Axel Huebl, Chad Mitchell
# License: BSD-3-Clause-LBNL
#
# -*- coding: utf-8 -*-
import sys
import tarfile
from urllib import request
import numpy as np
from surrogate_model_definitions import surrogate_model
try:
import torch
except ImportError:
print("Warning: Cannot import PyTorch. Skipping test.")
sys.exit(0)
from impactx import (
ImpactX,
ImpactXParIter,
RefPart,
TransformationDirection,
coordinate_transformation,
distribution,
elements,
)
def download_and_unzip(url, data_dir):
request.urlretrieve(url, data_dir)
with tarfile.open(data_dir) as tar_dataset:
tar_dataset.extractall()
# load models
N_stage = 9
data_url = (
"https://zenodo.org/records/10368972/files/ml_example_inference.tar.gz?download=1"
)
download_and_unzip(data_url, "inference_dataset")
dataset_dir = "datasets/"
model_dir = "models/"
model_list = [
surrogate_model(
dataset_dir + f"dataset_beam_stage_{i}.pt",
model_dir + f"beam_stage_{i}_model.pt",
)
for i in range(N_stage)
]
# information specific to the WarpX simulation
# for which the neural networks are surrogates
ebeam_lpa_z0 = -107e-6
L_plasma = 0.28
L_transport = 0.03
L_stage_period = L_plasma + L_transport
drift_after_LPA = 43e-6
L_surrogate = abs(ebeam_lpa_z0) + L_plasma + drift_after_LPA
# number of slices per ds in each lattice element
ns = 1
class LPASurrogateStage(elements.Programmable):
def __init__(self, stage_i, surrogate_model, surrogate_length, stage_start):
elements.Programmable.__init__(self)
self.stage_i = stage_i
self.surrogate_model = surrogate_model
self.surrogate_length = surrogate_length
self.stage_start = stage_start
self.push = self.surrogate_push
self.ds = surrogate_length
def surrogate_push(self, pc, step):
array = np.array
ref_part = pc.ref_particle()
ref_z_i = ref_part.z
ref_z_i_LPA = ref_z_i - self.stage_start
ref_z_f = ref_z_i + self.surrogate_length
ref_part_tensor = torch.tensor(
[ref_part.x, ref_part.y, ref_z_i_LPA, ref_part.px, ref_part.py, ref_part.pz]
)
ref_beta_gamma = np.sqrt(torch.sum(ref_part_tensor[3:] ** 2))
with torch.no_grad():
ref_part_model_final = self.surrogate_model(ref_part_tensor.float())
ref_uz_f = ref_part_model_final[5]
ref_beta_gamma_final = (
ref_uz_f # NOT np.sqrt(torch.sum(ref_part_model_final[3:]**2))
)
ref_part_final = torch.tensor([0, 0, ref_z_f, 0, 0, ref_uz_f])
# transform
coordinate_transformation(pc, TransformationDirection.to_fixed_t)
for lvl in range(pc.finest_level + 1):
for pti in ImpactXParIter(pc, level=lvl):
aos = pti.aos()
aos_arr = array(aos, copy=False)
soa = pti.soa()
real_arrays = soa.GetRealData()
px = array(real_arrays[0], copy=False)
py = array(real_arrays[1], copy=False)
pt = array(real_arrays[2], copy=False)
data_arr = (
torch.tensor(
np.vstack(
[aos_arr["x"], aos_arr["y"], aos_arr["z"], real_arrays[:3]]
)
)
.float()
.T
)
data_arr[:, 0] += ref_part.x
data_arr[:, 1] += ref_part.y
data_arr[:, 2] += ref_z_i_LPA
data_arr[:, 3:] *= ref_beta_gamma
data_arr[:, 3] += ref_part.px
data_arr[:, 4] += ref_part.py
data_arr[:, 5] += ref_part.pz
# # TODO this part needs to be corrected for general geometry
# # where the initial vector might not point in z
# # and even if it does, bending elements may change the direction
# # i.e. do we need to make sure beam is pointing in the right direction?
# # assume for now it is
with torch.no_grad():
data_arr_post_model = self.surrogate_model(data_arr.float())
# need to add stage start to z
data_arr_post_model[:, 2] += self.stage_start
# back to ref particle coordinates
for ii in range(3):
data_arr_post_model[:, ii] -= ref_part_final[ii]
data_arr_post_model[:, 3 + ii] -= ref_part_final[3 + ii]
data_arr_post_model[:, 3 + ii] /= ref_beta_gamma_final
aos_arr["x"] = data_arr_post_model[:, 0]
aos_arr["y"] = data_arr_post_model[:, 1]
aos_arr["z"] = data_arr_post_model[:, 2]
px[:] = data_arr_post_model[:, 3]
py[:] = data_arr_post_model[:, 4]
pt[:] = data_arr_post_model[:, 5]
# TODO this part needs to be corrected for general geometry
# where the initial vector might not point in z
# and even if it does, bending elements may change the direction
ref_part.x = ref_part_final[0]
ref_part.y = ref_part_final[1]
ref_part.z = ref_part_final[2]
ref_gamma = np.sqrt(1 + ref_beta_gamma_final**2)
ref_part.px = ref_part_final[3]
ref_part.py = ref_part_final[4]
ref_part.pz = ref_part_final[5]
ref_part.pt = -ref_gamma
# for now, I am applying the hack of manually setting s=z=ct.
# this will need to be revisited and evaluated more correctly
# when the accelerator length is more consistently established
ref_part.s += self.surrogate_length
ref_part.t += self.surrogate_length
# ref_part.s += pge1.ds
# ref_part.t += pge1.ds / ref_beta
coordinate_transformation(pc, TransformationDirection.to_fixed_s)
## Done!
lpa_stage_list = []
for i in range(N_stage):
lpa = LPASurrogateStage(i, model_list[i], L_surrogate, L_stage_period * i)
lpa.nslice = ns
lpa.ds = L_surrogate
lpa_stage_list.append(lpa)
#########
sim = ImpactX()
# set numerical parameters and IO control
sim.particle_shape = 2 # B-spline order
sim.space_charge = False
# sim.diagnostics = False # benchmarking
sim.slice_step_diagnostics = True
# domain decomposition & space charge mesh
sim.init_grids()
# load a 1 GeV electron beam with an initial
# normalized rms emittance of 1 nm
ref_u = 1957
energy_gamma = np.sqrt(1 + ref_u**2)
energy_MeV = 0.510998950 * energy_gamma # reference energy
bunch_charge_C = 10.0e-15 # used with space charge
npart = 10000 # number of macro particles
# reference particle
ref = sim.particle_container().ref_particle()
ref.set_charge_qe(-1.0).set_mass_MeV(0.510998950).set_kin_energy_MeV(energy_MeV)
ref.z = ebeam_lpa_z0
# particle bunch
distr = distribution.Gaussian(
sigmaX=0.75e-6,
sigmaY=0.75e-6,
sigmaT=0.1e-6,
sigmaPx=1.33 / energy_gamma,
sigmaPy=1.33 / energy_gamma,
sigmaPt=1e-8,
muxpx=0.0,
muypy=0.0,
mutpt=0.0,
)
sim.add_particles(bunch_charge_C, distr, npart)
pc = sim.particle_container()
L_transport = 0.03
L_lens = 0.003
L_focal = 0.5 * L_transport
L_drift = 0.5 * (L_transport - L_lens)
Kxy = np.sqrt(2.0 / L_focal / L_lens)
Kt = 1e-11
L_drift_minus_surrogate = L_drift
L_drift_1 = L_drift - drift_after_LPA
L_drift_before_2nd_stage = abs(ebeam_lpa_z0)
L_drift_2 = L_drift - L_drift_before_2nd_stage
#########
###
monitor = elements.BeamMonitor("monitor")
for i in range(N_stage):
sim.lattice.extend(
[
monitor,
lpa_stage_list[i],
]
)
if i != N_stage - 1:
sim.lattice.extend(
[
monitor,
elements.Drift(ds=L_drift_1),
monitor,
elements.ConstF(ds=L_lens, kx=Kxy, ky=Kxy, kt=Kt),
monitor,
elements.Drift(ds=L_drift_2),
]
)
sim.lattice.extend([monitor])
sim.evolve()
del sim
Analyze
We run the following script to analyze correctness:
Script analyze_ml_surrogate.py
#!/usr/bin/env python3
#
# Copyright 2022-2023 ImpactX contributors
# Authors: Ryan Sandberg, Axel Huebl, Chad Mitchell
# License: BSD-3-Clause-LBNL
#
# -*- coding: utf-8 -*-
import numpy as np
import openpmd_api as io
from scipy.stats import moment
def get_moments(beam):
"""Calculate standard deviations of beam position & momenta
and emittance values
Returns
-------
sigx, sigy, sigt, emittance_x, emittance_y, emittance_t
"""
sigx = moment(beam["position_x"], moment=2) ** 0.5 # variance -> std dev.
sigpx = moment(beam["momentum_x"], moment=2) ** 0.5
sigy = moment(beam["position_y"], moment=2) ** 0.5
sigpy = moment(beam["momentum_y"], moment=2) ** 0.5
sigt = moment(beam["position_t"], moment=2) ** 0.5
sigpt = moment(beam["momentum_t"], moment=2) ** 0.5
epstrms = beam.cov(ddof=0)
emittance_x = (
sigx**2 * sigpx**2 - epstrms["position_x"]["momentum_x"] ** 2
) ** 0.5
emittance_y = (
sigy**2 * sigpy**2 - epstrms["position_y"]["momentum_y"] ** 2
) ** 0.5
emittance_t = (
sigt**2 * sigpt**2 - epstrms["position_t"]["momentum_t"] ** 2
) ** 0.5
return (sigx, sigy, sigt, emittance_x, emittance_y, emittance_t)
# initial/final beam
series = io.Series("diags/openPMD/monitor.bp", io.Access.read_only)
last_step = list(series.iterations)[-1]
initial = series.iterations[1].particles["beam"].to_df()
final = series.iterations[last_step].particles["beam"].to_df()
# compare number of particles
num_particles = 10000
assert num_particles == len(initial)
assert num_particles == len(final)
print("Initial Beam:")
sigx, sigy, sigt, emittance_x, emittance_y, emittance_t = get_moments(initial)
print(f" sigx={sigx:e} sigy={sigy:e} sigt={sigt:e}")
print(
f" emittance_x={emittance_x:e} emittance_y={emittance_y:e} emittance_t={emittance_t:e}"
)
atol = 0.0 # ignored
rtol = num_particles**-0.5 # from random sampling of a smooth distribution
print(f" rtol={rtol} (ignored: atol~={atol})")
assert np.allclose(
[sigx, sigy, sigt, emittance_x, emittance_y],
[
7.488319e-07,
7.501963e-07,
9.996533e-08,
5.052374e-10,
5.130370e-10,
],
rtol=rtol,
atol=atol,
)
atol = 1.0e-6
print(f" atol~={atol}")
assert np.allclose([emittance_t], [0.0], atol=atol)
print("")
print("Final Beam:")
sigx, sigy, sigt, emittance_x, emittance_y, emittance_t = get_moments(final)
print(f" sigx={sigx:e} sigy={sigy:e} sigt={sigt:e}")
print(
f" emittance_x={emittance_x:e} emittance_y={emittance_y:e} emittance_t={emittance_t:e}"
)
atol = 0.0 # ignored
rtol = num_particles**-0.5 # from random sampling of a smooth distribution
print(f" rtol={rtol} (ignored: atol~={atol})")
assert np.allclose(
[sigx, sigy, sigt, emittance_x, emittance_y, emittance_t],
[
3.062763e-07,
2.873031e-07,
1.021142e-07,
9.090898e-12,
9.579053e-12,
2.834852e-11,
],
rtol=rtol,
atol=atol,
)
Visualize
You can run the following script to visualize the beam evolution over time:
Script visualize_ml_surrogate.py
#!/usr/bin/env python3
#
# Copyright 2022-2023 ImpactX contributors
# Authors: Ryan Sandberg, Axel Huebl, Chad Mitchell
# License: BSD-3-Clause-LBNL
#
# -*- coding: utf-8 -*-
import argparse
import glob
from matplotlib import pyplot as plt
import numpy as np
import openpmd_api as io
import pandas as pd
from scipy.constants import c, e, m_e
def read_all_files(file_pattern):
"""Read in all CSV files from each MPI rank (and potentially OpenMP
thread). Concatenate into one Pandas dataframe.
Returns
-------
pandas.DataFrame
"""
return pd.concat(
(
pd.read_csv(filename, delimiter=r"\s+")
for filename in glob.glob(file_pattern)
),
axis=0,
ignore_index=True,
).set_index("id")
def read_file(file_pattern):
for filename in glob.glob(file_pattern):
df = pd.read_csv(filename, delimiter=r"\s+")
if "step" not in df.columns:
step = int(re.findall(r"[0-9]+", filename)[0])
df["step"] = step
yield df
def read_time_series(file_pattern):
"""Read in all CSV files from each MPI rank (and potentially OpenMP
thread). Concatenate into one Pandas dataframe.
Returns
-------
pandas.DataFrame
"""
return pd.concat(
read_file(file_pattern),
axis=0,
ignore_index=True,
) # .set_index('id')
from enum import Enum
class TCoords(Enum):
REF = 1
GLOBAL = 2
def to_t(
ref_pz, ref_pt, data_arr_s, ref_z=None, coord_type=TCoords.REF
): # x, y, t, dpx, dpy, dpt):
"""Change to fixed t coordinates
Parameters
---
ref_pz: float, reference particle momentum in z
ref_pt: float, reference particle pt = -gamma
data_arr_s: Nx6 array-like structure containing fixed-s particle coordinates
ref_z: if transforming to global coordinates
coord_type: TCoords enum, (default is in ref coordinates) whether to get particle data relative to reference coordinate or in the global frame
Notes
"""
if type(data_arr_s) is pd.core.frame.DataFrame:
coordinate_columns = [
"position_x",
"position_y",
"position_t",
"momentum_x",
"momentum_y",
"momentum_t",
]
assert all(
val in data_arr_s.columns for val in coordinate_columns
), f"data_arr_s must have columns {' '.join(coordinate_columns)}"
x, y, t, dpx, dpy, dpt = data_arr_s[coordinate_columns].to_numpy().T
x = data_arr_s["position_x"]
y = data_arr_s["position_y"]
t = data_arr_s["position_t"]
dpx = data_arr_s["momentum_x"]
dpy = data_arr_s["momentum_y"]
dpt = data_arr_s["momentum_t"]
elif type(data_arr_s) is np.ndarray:
assert (
data_arr_s.shape[1] == 6
), f"data_arr_s.shape={data_arr_s.shape} but data_arr_s must be an Nx6 array"
x, y, t, dpx, dpy, dpt = data_arr_s.T
else:
raise Exception(
f"Incompatible input type {type(data_arr_s)} for data_arr_s, must be pandas DataFrame or Nx6 array-like object"
)
x += ref_pz * dpx * t / (ref_pt + ref_pz * dpt)
y += ref_pz * dpy * t / (ref_pt + ref_pz * dpt)
pz = np.sqrt(
-1 + (ref_pt + ref_pz * dpt) ** 2 - (ref_pz * dpx) ** 2 - (ref_pz * dpy) ** 2
)
t *= pz / (ref_pt + ref_pz * dpt)
if type(data_arr_s) is pd.core.frame.DataFrame:
data_arr_s["momentum_t"] = pz - ref_pz
dpt = data_arr_s["momentum_t"]
else:
dpt[:] = pz - ref_pz
if coord_type is TCoords.REF:
print("applying reference normalization")
dpt /= ref_pz
elif coord_type is TCoords.GLOBAL:
assert (
ref_z is not None
), "Reference particle z coordinate is required to transform to global coordinates"
print("target global coordinates")
t += ref_z
dpx *= ref_pz
dpy *= ref_pz
dpt += ref_pz
# data_arr_t = np.column_stack([xt,yt,z,dpx,dpy,dpz])
return # modifies data_arr_s in place
def plot_beam_df(
beam_at_step,
axT,
unit=1e6,
unit_z=1e3,
unit_label="$\mu$m",
unit_z_label="mm",
alpha=1.0,
cmap=None,
color="k",
size=0.1,
t_offset=0.0,
label=None,
z_ticks=None,
):
ax = axT[0][0]
ax.scatter(
beam_at_step.position_x.multiply(unit),
beam_at_step.position_y.multiply(unit),
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"x [%s]" % unit_label)
ax.set_ylabel(r"y [%s]" % unit_label)
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
###########
ax = axT[0][1]
ax.scatter(
beam_at_step.position_t.multiply(unit_z) - t_offset,
beam_at_step.position_x.multiply(unit),
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"%s" % unit_z_label)
ax.set_ylabel(r"x [%s]" % unit_label)
ax.axes.ticklabel_format(axis="y", style="sci", scilimits=(-2, 2))
if z_ticks is not None:
ax.set_xticks(z_ticks)
###########
ax = axT[0][2]
ax.scatter(
beam_at_step.position_t.multiply(unit_z) - t_offset,
beam_at_step.position_y.multiply(unit),
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"%s" % unit_z_label)
ax.set_ylabel(r"y [%s]" % unit_label)
ax.axes.ticklabel_format(axis="y", style="sci", scilimits=(-2, 2))
if z_ticks is not None:
ax.set_xticks(z_ticks)
############
##########
ax = axT[1][0]
ax.scatter(
beam_at_step.momentum_x,
beam_at_step.momentum_y,
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel("px")
ax.set_ylabel("py")
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
##########
ax = axT[1][1]
ax.scatter(
beam_at_step.momentum_t,
# beam_at_step.position_t.multiply(unit_z)-t_offset,
beam_at_step.momentum_x,
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel("pt")
# ax.set_xlabel(r'%s'%unit_z_label)
ax.set_ylabel("px")
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
##########
ax = axT[1][2]
ax.scatter(
beam_at_step.momentum_t,
# beam_at_step.position_t.multiply(unit_z)-t_offset,
beam_at_step.momentum_y,
c=color,
s=size,
alpha=alpha,
label=label,
cmap=cmap,
)
if label is not None:
ax.legend()
# ax.set_xlabel(r'%s'%unit_z_label)
ax.set_xlabel("pt")
ax.set_ylabel("py")
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
############
############
##########
ax = axT[2][0]
ax.scatter(
beam_at_step.position_x.multiply(unit),
beam_at_step.momentum_x,
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"x [%s]" % unit_label)
ax.set_ylabel("px")
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
############
ax = axT[2][1]
ax.scatter(
beam_at_step.position_y.multiply(unit),
beam_at_step.momentum_y,
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"y [%s]" % unit_label)
ax.set_ylabel("py")
ax.axes.ticklabel_format(axis="both", style="sci", scilimits=(-2, 2))
################
ax = axT[2][2]
ax.scatter(
beam_at_step.position_t.multiply(unit_z) - t_offset,
beam_at_step.momentum_t,
c=color,
s=size,
alpha=alpha,
cmap=cmap,
)
ax.set_xlabel(r"%s" % unit_z_label)
ax.set_ylabel("pt")
ax.axes.ticklabel_format(axis="y", style="sci", scilimits=(-2, 2))
if z_ticks is not None:
ax.set_xticks(z_ticks)
plt.tight_layout()
# done
# options to run this script
parser = argparse.ArgumentParser(description="Plot the ML surrogate benchmark.")
parser.add_argument(
"--save-png", action="store_true", help="non-interactive run: save to PNGs"
)
args = parser.parse_args()
impactx_surrogate_reduced_diags = read_time_series(
"diags/reduced_beam_characteristics.*"
)
ref_gamma = np.sqrt(1 + impactx_surrogate_reduced_diags["ref_beta_gamma"] ** 2)
beam_gamma = (
ref_gamma
- impactx_surrogate_reduced_diags["pt_mean"]
* impactx_surrogate_reduced_diags["ref_beta_gamma"]
)
beam_u = np.sqrt(beam_gamma**2 - 1)
emit_x = impactx_surrogate_reduced_diags["emittance_x"]
emit_nx = emit_x * beam_u
emit_y = impactx_surrogate_reduced_diags["emittance_y"]
emit_ny = emit_y * beam_u
ix_slice = [0] + [2 + 9 * i for i in range(8)]
############# plot moments ##############
fig, axT = plt.subplots(2, 2, figsize=(10, 8))
######### emittance ##########
ax = axT[0][0]
scale = 1e6
ax.plot(
impactx_surrogate_reduced_diags["s"][ix_slice],
emit_nx[ix_slice] * scale,
"bo",
label="x",
)
ax.plot(
impactx_surrogate_reduced_diags["s"][ix_slice],
emit_ny[ix_slice] * scale,
"ro",
label="y",
)
ax.legend()
ax.set_xlabel("s [m]")
ax.set_ylabel(r"emittance (mm-mrad)")
######### energy ##########
ax = axT[0][1]
scale = m_e * c**2 / e * 1e-9
ax.plot(
impactx_surrogate_reduced_diags["s"][ix_slice],
beam_gamma[ix_slice] * scale,
"go",
)
ax.set_xlabel("s [m]")
ax.set_ylabel(r"mean energy (GeV)")
######### width ##########
ax = axT[1][0]
scale = 1e6
ax.plot(
impactx_surrogate_reduced_diags["s"][ix_slice],
impactx_surrogate_reduced_diags["sig_x"][ix_slice] * scale,
"bo",
label="x",
)
ax.plot(
impactx_surrogate_reduced_diags["s"][ix_slice],
impactx_surrogate_reduced_diags["sig_y"][ix_slice] * scale,
"ro",
label="y",
)
ax.legend()
ax.set_xlabel("s [m]")
ax.set_ylabel(r"beam width ($\mu$m)")
######### divergence ##########
ax = axT[1][1]
scale = 1e3
ax.semilogy(
impactx_surrogate_reduced_diags["s"][ix_slice],
impactx_surrogate_reduced_diags["sig_px"][ix_slice] * scale,
"bo",
label="x",
)
ax.semilogy(
impactx_surrogate_reduced_diags["s"][ix_slice],
impactx_surrogate_reduced_diags["sig_py"][ix_slice] * scale,
"ro",
label="y",
)
ax.legend()
ax.set_xlabel("s [m]")
ax.set_ylabel(r"divergence (mrad)")
plt.tight_layout()
if args.save_png:
plt.savefig("lpa_ml_surrogate_moments.png")
else:
plt.show()
######## plot phase spaces ###########
beam_impactx_surrogate_series = io.Series(
"diags/openPMD/monitor.bp", io.Access.read_only
)
impactx_surrogate_steps = list(beam_impactx_surrogate_series.iterations)
impactx_surrogate_ref_particle = read_time_series("diags/ref_particle.*")
millimeter = 1.0e3
micron = 1.0e6
N_stage = 9
impactx_stage_end_steps = [1] + [3 + 8 * i for i in range(N_stage)]
ise = impactx_stage_end_steps
# initial
step = 1
beam_at_step = beam_impactx_surrogate_series.iterations[step].particles["beam"].to_df()
ref_part_step = impactx_surrogate_ref_particle.loc[step]
ref_u = np.sqrt(ref_part_step["pt"] ** 2 - 1)
to_t(
ref_u,
ref_part_step["pt"],
beam_at_step,
ref_z=ref_part_step["z"],
coord_type=TCoords.GLOBAL,
)
t_offset = impactx_surrogate_ref_particle.loc[step, "t"] * micron
fig, axT = plt.subplots(3, 3, figsize=(10, 8))
fig.suptitle(f"initially, ct={impactx_surrogate_ref_particle.at[step,'t']:.2e}")
plot_beam_df(
beam_at_step,
axT,
alpha=0.6,
color="red",
unit_z=1e6,
unit_z_label=r"$\xi$ [$\mu$m]",
t_offset=t_offset,
z_ticks=[-107.3, -106.6],
)
if args.save_png:
plt.savefig(f"initial_phase_spaces.png")
else:
plt.show()
####### final ###########
stage_i = 8
step = ise[stage_i + 1]
beam_at_step = beam_impactx_surrogate_series.iterations[step].particles["beam"].to_df()
ref_part_step = impactx_surrogate_ref_particle.loc[step]
ref_u = np.sqrt(ref_part_step["pt"] ** 2 - 1)
to_t(
ref_u,
ref_part_step["pt"],
beam_at_step,
ref_z=ref_part_step["z"],
coord_type=TCoords.GLOBAL,
)
t_offset = impactx_surrogate_ref_particle.loc[step, "t"] * micron
fig, axT = plt.subplots(3, 3, figsize=(10, 8))
fig.suptitle(f"stage {stage_i}, ct={impactx_surrogate_ref_particle.at[step,'t']:.2e}")
plot_beam_df(
beam_at_step,
axT,
alpha=0.6,
color="red",
unit_z=1e6,
unit_z_label=r"$\xi$ [$\mu$m]",
t_offset=t_offset,
z_ticks=[-107.3, -106.6],
)
if args.save_png:
plt.savefig(f"stage_{stage_i}_phase_spaces.png")
else:
plt.show()
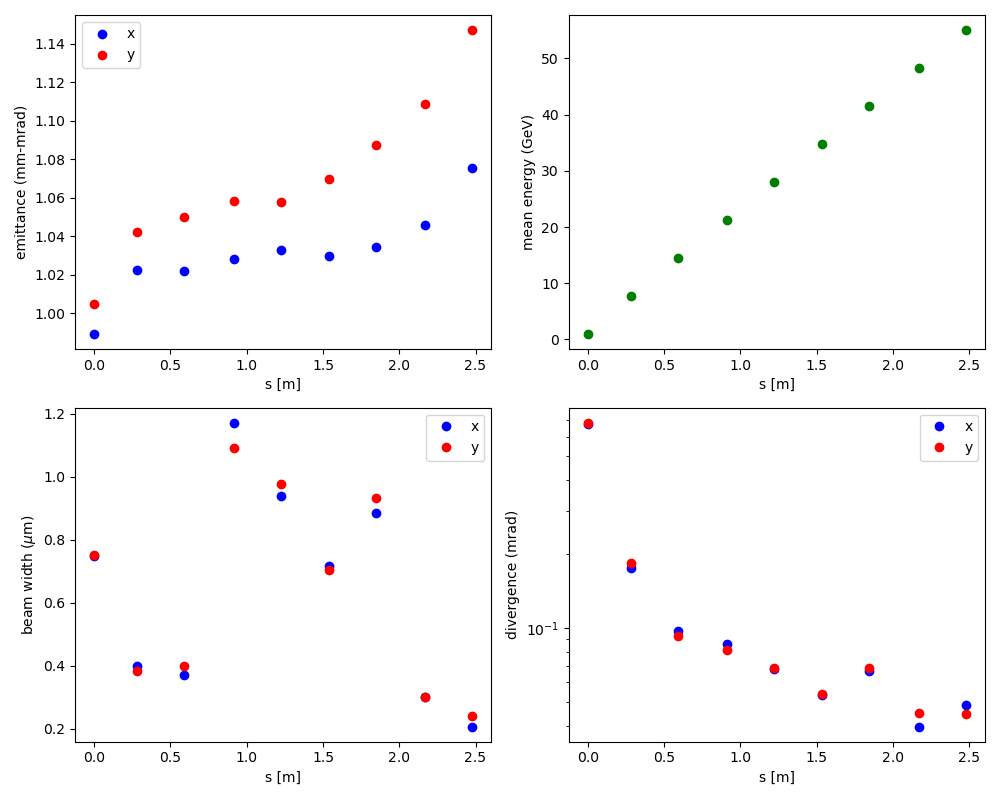
Fig. 7 Evolution of electron beam moments through 9 stages of LPAs (via neural network surrogates).

Fig. 8 Initial phase space projections going into 9 stage LPA (via neural network surrogates) simulation. Top row: spatial projections, middle row: momentum projections, bottom row: phase spaces.
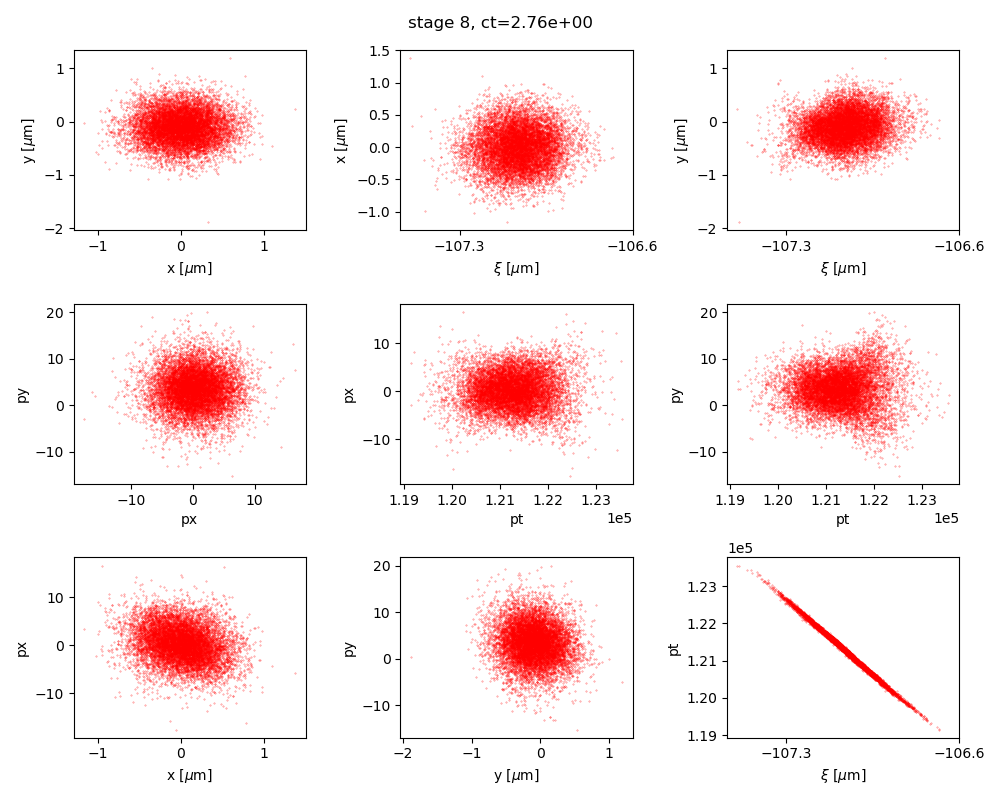
Fig. 9 Final phase space projections after 9 stage LPA (via neural network surrogates) simulation. Top row: spatial projections, middle row: momentum projections, bottom row: phase spaces.